Variational Autoencoders
Sources:
- VAE paper
- Mathematics Behind Variational AutoEncoders
- Variational autoencoders by Matthew N. Bernstein
- From Autoencoder to Beta-VAE
- Stanford CS 228: The variational auto-encoder
Link: My VAE implementation on Github
Notations
| Symbol | Mean |
|---|---|
| \(\mathcal{D}\) | The dataset, \(\mathcal{D} = \left\{ \mathbf{x}^{(1)}, \mathbf{x}^{(2)}, \ldots, \mathbf{x}^{(n)} \right\}\), contains \(n\) data samples; \(|\mathcal{D}| = n\). |
| \(\mathbf{x}^{(i)}\) | Each data point is a vector of \(d\) dimensions, \(\mathbf{x}^{(i)} = \left[ x_1^{(i)}, x_2^{(i)}, \ldots, x_d^{(i)} \right]\). |
| \(\mathbf{x}\) | One data sample from the dataset, \(\mathbf{x} \in \mathcal{D}\). |
| \(\mathbf{x}^{\prime}\) | The reconstructed version of \(\mathbf{x}\). |
| \(\tilde{\mathbf{x}}\) | The corrupted version of \(\mathbf{x}\). |
| \(\mathbf{z}\) | The compressed code learned in the bottleneck layer. |
| \(a_j^{(l)}\) | The activation function for the \(j\)-th neuron in the \(l\)-th hidden layer. |
| \(g_\phi(\cdot)\) | The encoding function parameterized by \(\phi\). |
| \(f_\theta(\cdot)\) | The decoding function parameterized by \(\theta\). |
| \(q_\phi(\mathbf{z} \mid \mathbf{x})\) | Estimated posterior probability function, also known as probabilistic encoder. |
| \(p_\theta(\mathbf{x} \mid \mathbf{z})\) | Likelihood of generating true data sample given the latent code, also known as probabilistic decoder. d |
Autoencoders
Autoencoder is a neural network designed to learn an identity function in an unsupervised way to reconstruct the original input while compressing the data in the process so as to discover a more efficient and compressed representation. The idea was originated in the 1980s, and later promoted by the seminal paper by Hinton & Salakhutdinov, 2006.
It consists of two networks:
- Encoder network: It translates the original high-dimension input into the latent low-dimensional code. The input size is larger than the output size.
- Decoder network: The decoder network recovers the data from the code, likely with larger and larger output layers.
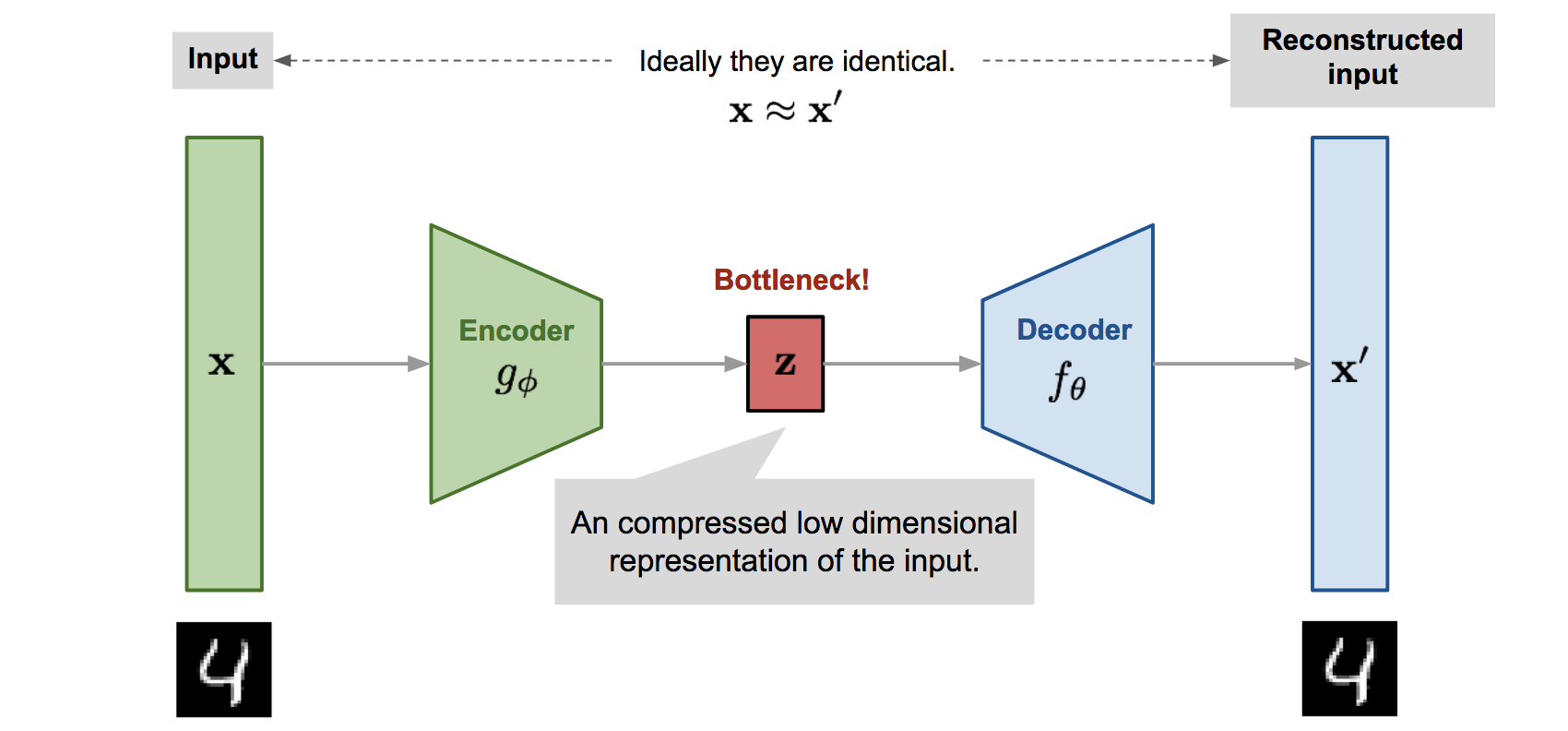
The encoder network essentially accomplishes the dimensionality reduction, just like how we would use Principal Component Analysis (PCA) or Matrix Factorization (MF) for
The model contains:
- an encoder function \(g(.)\) parameterized by \(\phi\) and
- a decoder function \(f(.)\) parameterized by \(\theta\).
The low-dimensional code learned for input \(\mathbf{x}\) in the bottleneck layer is \(\mathbf{z}=g_\phi(\mathbf{x})\) and the reconstructed input is \(\mathbf{x}^{\prime}=f_\theta\left(g_\phi(\mathbf{x})\right)\).
The parameters \((\theta, \phi)\) are learned together to output a reconstructed data sample same as the original input, \(\mathbf{x} \approx f_\theta\left(g_\phi(\mathbf{x})\right)\), or in other words, to learn an identity function.
There are various metrics to quantify the difference between two vectors, such as cross entropy when the activation function is sigmoid, or as simple as MSE loss: \[ L_{\mathrm{AE}}(\theta, \phi)=\frac{1}{n} \sum_{i=1}^n\left(\mathbf{x}^{(i)}-f_\theta\left(g_\phi\left(\mathbf{x}^{(i)}\right)\right)\right)^2 \]
VAE: Variational Autoencoder
The idea of Variational Autoencoder (Kingma & Welling, 2014), short for VAE, is actually less similar to all the autoencoder models above, but deeply rooted in the methods of variational bayesian and graphical model.
Let's consider some dataset consisting of \(\mathrm{N}\) i.i.d. samples of a random variable \(\mathbf{x}\) (either scalar or vector) (Q: why iid?). The data are assumed to be generated by some random process, involving an unobserved random variable \(\mathbf{z}\) (i.e. the latent variable).
The generative process has two steps: 1. a value \(\mathbf{z}^{(\mathbf{i})}\) is generated from some prior distribution \(p_{\boldsymbol{\theta}^*}(\mathbf{z})\), 2. a value \(\mathbf{x}^{(\mathbf{i})}\) is generated from some conditional distribution \(p_{\boldsymbol{\theta}^*}(\mathbf{x} \mid \mathbf{z} = \mathbf{z}^{(\mathbf{i})})\) dependent on \(\mathbf{z}^{(\mathbf{i})}\),
We assume that the prior \(p_{\boldsymbol{\theta}^*}(\mathbf{z})\) and likelihood \(p_{\boldsymbol{\theta}^*}(\mathbf{x} \mid \mathbf{z})\) come from parametric families of distributions \(p_{\boldsymbol{\theta}}(\mathbf{z})\) and \(p_{\boldsymbol{\theta}}(\mathbf{x} \mid \mathbf{z})\), and that their PDFs are differentiable almost almost everywhere w.r.t. both \(θ\) and \(\mathbf{z}\).
(Q: Why we assume they both controled by \(\theta\)?)
The variational method
We are interested in \(p(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)})\), the posterior inference of the latent variable \(\mathbf{z}\) given an observed value \(\mathbf{x}\) for a choice of parameters \(\theta\). It is useful for representation learning.
We use the variational method to calculate it. The process is:
$$ \[\begin{aligned} p_\theta\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right) & = \frac{p_\theta\left(\mathbf{z}, \mathbf{x} = \mathbf{x}^{(i)}\right)}{p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)}\right)} \\ & = \frac { \textcolor{purple} {p_\theta\left(\mathbf{x}=\mathbf{x}^{(i)} \mid \mathbf{z}\right) p_\theta\left(\mathbf{z}\right)} } { \int_{\mathbf{z}^{(i)}} p_\theta \left(\mathbf{x}=\mathbf{x}^{(i)} \mid \mathbf{z}=\mathbf{z}^{(i)}\right) p_\theta\left(\mathbf{z}=\mathbf{z}^{(i)}\right) d \mathbf{z}^{(i)} } \end{aligned}\]$$
Suppose that we have a choice for parameters \(\theta\), thus both the specification of the prior distribution \(\textcolor{purple} {p_\theta(\mathbf{z})}\) and the likelihood \(\textcolor{purple} {p_\theta\left(\mathbf{x} =\mathbf{x}^{(\mathbf{i})} \mid \mathbf{z} \right)}\) defined by the generative process would be known.
Therefore theoretically, the posterior \(p_\theta\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(\mathbf{i})} \right)\) can be calculated after evaluating the integral in the denominator, which involves enumerating all the possible values the unobservable variable \(\mathrm{z}\) may have.
However without any simplifying assumptions on \(p\left(\mathbf{z} \mid \mathbf{x}\mathbf{x}^{(\mathbf{i})} ; \theta\right)\) or \(p(\mathbf{z} ; \theta)\), the integral is intractable, as it is very expensive to check all the possible values of \(\mathbf{z}\) and sum them up.
To solve this, the variational method introduces a so-called recognition model \(q_\phi(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)})\), parameterized by \(\phi\), as an approximation to the true posterior \(p_\theta\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)\).
Assumptions
In practice, we will always assume the encoder output distribution \(q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)\) to be a multivariate gaussian: \[ \mathbf{z} \sim q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right) = \mathcal{N}\left(\boldsymbol{\mu}^{(i)}, (\boldsymbol{\sigma}^{(i)})^2 \boldsymbol{I}\right) . \]
In practice, we will always assume the prior distribution \(p_\theta(z)\) to be a multivariate standard gaussian: \[ \mathbf{z} \sim p_\phi\left(\mathbf{z}\right)=\mathcal{N}\left(\boldsymbol{0}, \boldsymbol{I}\right) . \]
The loss function: negative ELBO
The estimated posterior \(q_\phi(\mathbf{z} \mid \mathbf{x})\) should be very close to the real one \(p_\theta(\mathbf{z} \mid \mathbf{x})\). We want to reduce the Kullback-Leibler divergence of between these two distributions: \[ \begin{aligned} D_{\mathrm{KL}}\left(q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right) \| p_\theta\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)\right) & = \int_{\mathbf{z}} q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right) \log \frac{q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)}{p_\theta\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)} d \mathbf{z} \\ & = \mathbb{E}_q\left[\log q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)\right] - \mathbb{E}_q\left[\log p_\theta\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)\right] \\ & = \mathbb{E}_q\left[\log q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)\right] - \mathbb{E}_q\left[\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)}, \mathbf{z}\right)\right] + \mathbb{E}_{q_\phi}\left[\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)}\right)\right] \\ & = \mathbb{E}_q\left[\log q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)\right] - \mathbb{E}_q\left[\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)}, \mathbf{z}\right)\right] + \log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)}\right) \\ & = -\text{ELBO}_\theta + \textcolor{blue}{\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)}\right) } \end{aligned} \]
(\(\mathbb{E}_q\) means \(\mathbb{E}_{q_{\phi}}\))
where ELBO stands for Evidence Lower BOund, which is defind as: \[ \begin{aligned} \mathrm{ELBO} & = -\mathbb{E}_q\left[\log q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)\right] + \mathbb{E}_q\left[\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)}, \mathbf{z}\right)\right] \\ & = -\mathbb{E}_q\left[\log q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right)\right] + \mathbb{E}_q\left[\log p_\theta\left(\mathbf{z}\right)\right] + \mathbb{E}_q\left[\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)} \mid \mathbf{z}\right)\right] \\ & = -D_{\mathrm{KL}}\left(q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right) \| p_\theta(\mathbf{z})\right) + \mathbb{E}_q\left[\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)} \mid \mathbf{z}\right)\right] \end{aligned} \] Since the term \(\textcolor{blue}{\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)}\right) }\) is a constant, it can be ignored during the optimization process. As the result, the loss function is: \[ \begin{aligned} \mathcal{L}\left(\phi, \theta, \mathbf{x}^{(\mathbf{i})}\right) & = - \mathrm{ELBO} \\ & = \textcolor{pink} {D_{\mathrm{KL}}\left(q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right) \| p_\theta(\mathbf{z})\right)} - \textcolor{green} {\mathbb{E}_q\left[\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)} \mid \mathbf{z}\right)\right] } \end{aligned} \] ## How to compute the loss function
The expectation term
We will use Monte Carlo method to calculate (or, more precisely, approximate) the expectation term, i.e., \[ \textcolor{green} {\mathbb{E}_q\left[\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)} \mid \mathbf{z}\right)\right] \approx \frac{1}{L} \sum_{l=1}^L\left[\log p\left(\mathbf{x}^{(i)} \mid \mathbf{z}^{(i l)} ; \theta\right)\right]} \] when \(L\) is very large.
However, in practice, we will set \(L=1\) to calculate the mean \(\textcolor{green} {\mathbb{E}_q\left[\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)} \mid \mathbf{z}\right)\right]}\) (Yeah, you are right. We only use one sample to approximate the expectation. That's radiculous! But somehow they just regulated it.)
So that our approximated mean equals \(\textcolor{green} {\log p\left(\mathbf{x}^{(i)} \mid \mathbf{z}\right)}\): \[ \textcolor{green} {\mathbb{E}_q\left[\log p_\theta\left(\mathbf{x} = \mathbf{x}^{(i)} \mid \mathbf{z}\right)\right] \approx \text{(very weird)} \quad \log p\left(\mathbf{x}^{(i)} \mid \mathbf{z}\right)} \]
The KL term
Meanwhile, according to our assumptions (see the previous section), we will always assume \(q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right) = \mathcal{N}\left(\boldsymbol{\mu}^{(i)}, (\boldsymbol{\sigma}^{(i)})^2 \boldsymbol{I}\right)\), \(p_\phi\left(\mathbf{z}\right)=\mathcal{N}\left(\boldsymbol{0}, \boldsymbol{I}\right)\). In this particular case, it turns out that the KL-divergence term above can be expressed analytically (See the Appendix to this post): \[ \textcolor{pink} {D_{\mathrm{KL}}\left(q_\phi\left(\mathbf{z} \mid \mathbf{x} = \mathbf{x}^{(i)}\right) \| p_\theta(\mathbf{z})\right)=-\frac{1}{2} \sum_{j=1}^J\left(1+\log \sigma_j^2-\mu_j^2-\sigma_j^2\right)} \]
The result
So that \[ \begin{aligned} \mathcal{L}\left(\phi, \theta, \mathbf{x}^{(\mathbf{i})}\right) & = \textcolor{pink} { \frac{1}{2} \sum_{j=1}^J\left(1+ \log \sigma^2_j-\mu_j^2-\sigma_j^2\right) } - \textcolor{green} {\log p\left(\mathbf{x}^{(i)} \mid \mathbf{z}\right)} . \end{aligned} \]
Why use reversed KL?
Back in the last section, The loss function: negative ELBO, we regulate the loss function to be \(D_{\mathrm{KL}}\left(q_\phi \mid p_\theta\right)\). But why we choose \(D_{\mathrm{KL}}\left(q_\phi \mid p_\theta\right)\) (reversed KL) instead of \(D_{\mathrm{KL}}\left(p_\theta \mid q_\phi\right)\) (forward KL)?
Eric Jang has a great explanation in his post on Bayesian Variational methods. As a quick recap:
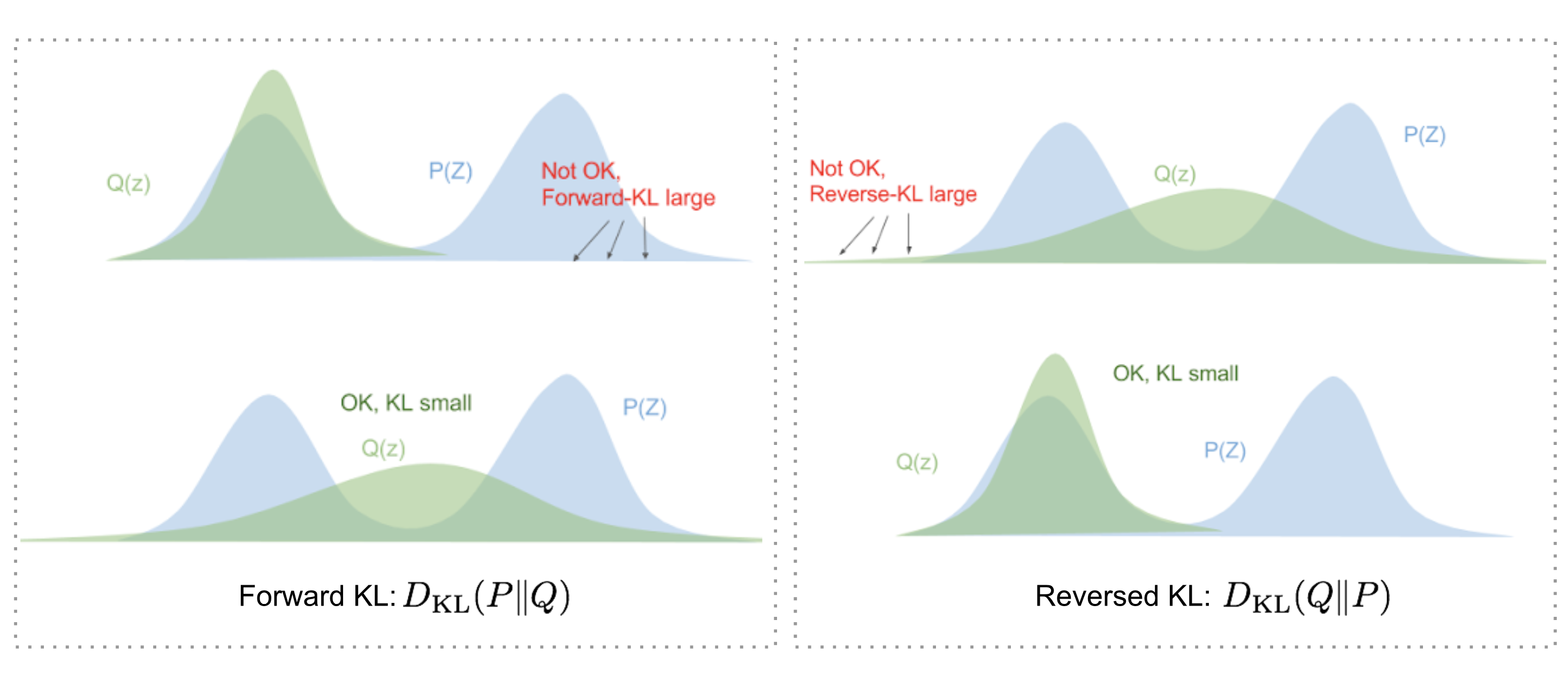
- Forward KL divergence: \(D_{\mathrm{KL}}(P \mid Q)=\mathbb{E}_{z \sim P(z)} \log \frac{P(z)}{Q(z)}\); we have to ensure that \(\mathrm{Q}(\mathrm{z})>0\) wherever \(\mathrm{P}(\mathrm{z})>0\). The optimized variational distribution \(q(z)\) has to cover over the entire \(p(z)\).
- Reversed KL divergence: \(D_{\mathrm{KL}}(Q \mid P)=\mathbb{E}_{z \sim Q(z)} \log \frac{Q(z)}{P(z)}\); minimizing the reversed KL divergence squeezes the \(Q(z)\) under \(P(z)\).
Tricks
Reparameterization trick
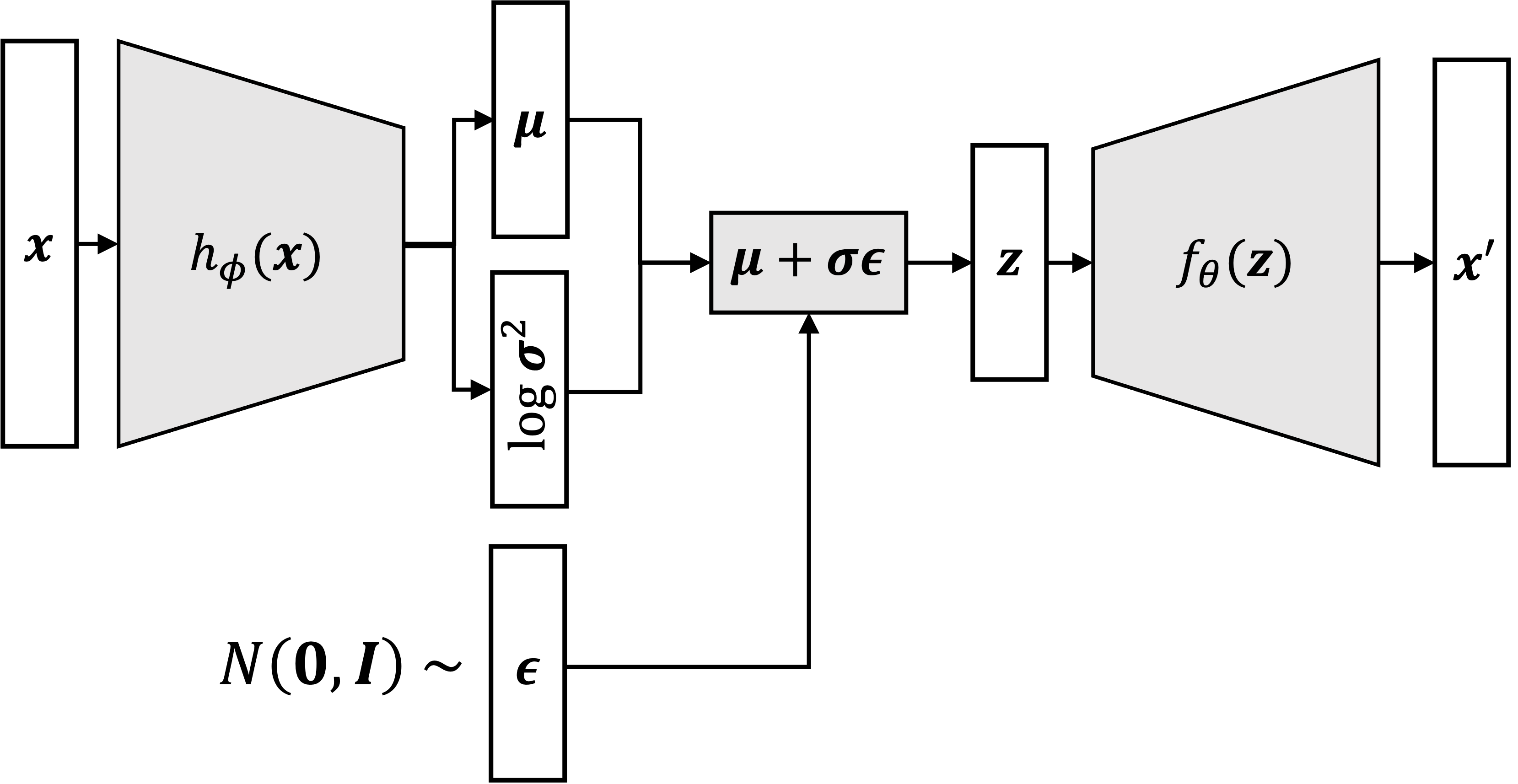
The expectation term in the loss function invokes generating samples from \(\mathbf{z} \sim q_\phi(\mathbf{z} \mid \mathbf{x})\). Sampling is a stochastic process and therefore we cannot backpropagate the gradient.
To make it trainable, the reparameterization trick is introduced: It is often possible to express the random variable \(\mathbf{z}\) as a deterministic variable \(\mathbf{z}=\mathcal{T}_\phi(\mathbf{x}, \boldsymbol{\epsilon})\), where \(\boldsymbol{\epsilon}\) is an auxiliary independent random variable, and the transformation function \(\mathcal{T}_\phi\) parameterized by \(\phi\) converts \(\epsilon\) to \(\mathbf{z}\).
For example, a common choice of the form of \(q_\phi(\mathbf{z} \mid \mathbf{x})\) is a multivariate Gaussian with a diagonal covariance structure: \[ \begin{aligned} & \mathbf{z} \sim q_\phi\left(\mathbf{z} \mid \mathbf{x}^{(i)}\right)=\mathcal{N}\left(\mathbf{z} ; \boldsymbol{\mu}^{(i)}, \boldsymbol{\sigma}^{2(i)} \boldsymbol{I}\right) \quad \text {; Assume it's Gaussian. } \\ & \mathbf{z}=\boldsymbol{\mu}+\boldsymbol{\sigma} \odot \boldsymbol{\epsilon}, \text { where } \boldsymbol{\epsilon} \sim \mathcal{N}(0, \boldsymbol{I}) \quad \text {; Reparameterization trick. } \end{aligned} \] where \(\odot\) refers to element-wise product.
1 | def reparameterization(self, mean, std_var): |
Log var trick
Instead of using variance \(\boldsymbol{\sigma}\), we use the log-variance to allow for positive and negative values: \(\log \left(\boldsymbol{\sigma}^2\right)\)
Why can we do this? \[ \begin{aligned} \log \left(\boldsymbol{\sigma}^2\right) & =2 \cdot \log (\boldsymbol{\sigma}) \\ \log \left(\boldsymbol{\sigma}^2\right) / 2 & =\log (\boldsymbol{\sigma}) \\ \boldsymbol{\sigma} & =e^{\log \left(\boldsymbol{\sigma}^2\right) / 2} \end{aligned} \]
So, when we sample the points, we can do \[ \mathbf{z}=\boldsymbol{\mu}+e^{\log \left(\boldsymbol{\sigma}^2\right) / 2} \odot \boldsymbol{\epsilon} . \]
Model archiecture
VAE model
1 | class Model(nn.Module): |
Encoder
1 | """ |
Decoder
1 | class Decoder(nn.Module): |
Training process
1 | import torch |
Hyperparameters:
1 | # Model Hyperparameters |
Load the dataset
1 | from torchvision.datasets import MNIST |
Build the model
1 | encoder = Encoder(input_dim=x_dim, hidden_dim=hidden_dim, latent_dim=latent_dim) |
Define the loss function
1 | def loss_function(x, x_hat, mean, log_var): |
Start training loop
1 | print("Start training VAE...") |
Appendix
Derivation of the KL-divergence term when the variational posterior and prior are Gaussian
If we choose the variational distribution \(q_\phi(\boldsymbol{z} \mid \boldsymbol{x})\) to be defined by the following generative process: \[ \begin{aligned} \boldsymbol{\mu} & :=h_\phi^{(1)}(\boldsymbol{x}) \\ \log \boldsymbol{\sigma}^2 & :=h_\phi^{(2)}(\boldsymbol{x}) \\ \boldsymbol{z} & \sim N\left(\boldsymbol{\mu}, \boldsymbol{\sigma}^{\mathbf{2}} \boldsymbol{I}\right) \end{aligned} \] where \(h^{(1)}\) and \(h^{(2)}\) are the two encoding neural networks, and we choose the prior distribution over \(z\) to be a standard normal distribution: \[ p(z):=N(\mathbf{0}, \boldsymbol{I}) \] then the KL-divergence from \(p(z)\) to \(q_\phi(\boldsymbol{z} \mid \boldsymbol{x})\) is given by the following formula: \[ D_{\mathrm{KL}}\left(q_\phi(\boldsymbol{z} \mid \boldsymbol{x}) \| p(z)\right)=-\frac{1}{2} \sum_{j=1}^J\left(1+h_\phi^{(2)}(\boldsymbol{x})_j-\left(h_\phi^{(1)}(\boldsymbol{x})\right)_j^2-\exp \left(h_\phi^2(\boldsymbol{x})_j\right)\right) \] where \(J\) is the dimensionality of \(z\).
Proof:
First, let's re-write the KL-divergence as follows: \[ \begin{align*} D_{\mathrm{KL}}(q_\phi(\boldsymbol{z} \mid \boldsymbol{x}) || p(\boldsymbol{z})) &= \int q_\phi(\boldsymbol{z} \mid \boldsymbol{x}) \log \frac{q_\phi(\boldsymbol{z} \mid \boldsymbol{x})}{ p(\boldsymbol{z}))} \\ &= \int q_\phi(\boldsymbol{z} \mid \boldsymbol{x}) \log q_\phi(\boldsymbol{z} \mid \boldsymbol{x}) \ d\boldsymbol{z} - \int q_\phi(\boldsymbol{z} \mid \boldsymbol{x}) \log p(\boldsymbol{z}) \ d\boldsymbol{z}\end{align*} \]
First, let's compute the first term, \(\int q_\phi(\boldsymbol{z} \mid \boldsymbol{x}) \log q_\phi(\boldsymbol{z} \mid \boldsymbol{x}) d z\) : \[ \begin{aligned} \int q_\phi(z \mid x) \log q_\phi(z \mid x) d z & =\int N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) \log N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) d z \\ & =\int N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) \sum_{j=1}^J \log N\left(z_j ; \mu_j, \sigma_j^2\right) d z \\ & =\int N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) \sum_{j=1}^J\left[\log \left(\frac{1}{\sqrt{\sigma^2 2 \pi}}\right)-\frac{1}{2} \frac{\left(z_j-\mu_j\right)^2}{\sigma_j^2}\right] d z \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J \log \sigma_j^2-\frac{1}{2} \sum_{j=1}^J \int N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) \frac{\left(z_j-\mu_j\right)^2}{\sigma_j^2} d z \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J \log \sigma_j^2-\frac{1}{2} \sum_{j=1}^J \int_{z_j} N\left(z_j ; \mu_j, \sigma_j^2\right) \frac{\left(z_j-\mu_j\right)^2}{\sigma_j^2} \int_{z_{i \neq j}} \prod_{i \neq j} N\left(z_i ; \mu_i, \sigma_i^2\right) d z_{i \neq j} d z_j \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J \log \sigma_j^2-\frac{1}{2} \sum_{j=1}^J \int N\left(z_j ; \mu_j, \sigma_j^2\right) \frac{\left(z_j-\mu_j\right)^2}{\sigma_j^2} d z_j \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J \log \sigma_j^2-\frac{1}{2} \sum_{j=1}^J \frac{1}{\sigma_j^2} \int N\left(z_j ; \mu_j, \sigma_j^2\right)\left(z_j^2-2 z_j \mu_j+\mu_j^2\right) d z_j \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J \log \sigma_j^2-\frac{1}{2} \sum_{j=1}^J \frac{1}{\sigma_j^2}\left(E\left[z_j^2\right]-E\left[2 z_j \mu_j\right]+\mu_j^2\right) \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J \log \sigma_j^2-\frac{1}{2} \sum_{j=1}^J \frac{1}{\sigma_j^2}\left(\mu_j^2+\sigma^2-2 \mu_j^2+\mu_j^2\right) \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J \log \sigma_j^2-\frac{1}{2} \sum_{j=1}^J 1 \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J\left(1+\log \sigma_j^2\right) \end{aligned} \]
Now, let us compute the second term, \(\int q_\phi(\boldsymbol{z} \mid \boldsymbol{x}) \log p(z) d z\) : \[ \begin{aligned} \int q_\phi(z \mid x) \log p(z) d z & \left.=\int N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) \log N(z ; \mathbf{0}, \boldsymbol{I})\right) d z \\ & =\int N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) \sum_{j=1}^J N\left(z_i ; 0,1\right) d z \\ & =\int N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) \sum_{j=1}^J\left[\log \frac{1}{\sqrt{2 \pi}}-\frac{1}{2} z_i^2\right] d z \\ & =J \log \frac{1}{\sqrt{2 \pi}} \int N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) d z-\frac{1}{2} \int N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) \sum_{j=1}^J z_i^2 d z \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \int N\left(z ; \boldsymbol{\mu}, \operatorname{diag}\left(\boldsymbol{\sigma}^2\right)\right) \sum_{j=1}^J z_i^2 d z \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \int \sum_{j=1}^J z_j^2 \prod_{j^{\prime}=1}^J N\left(z_{j^{\prime}} ; \mu_{j^{\prime}}, \sigma_{j^{\prime}}^2\right) d z \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J \int_{z_j} z_j^2 N\left(z_j ; \mu_j, \sigma_j^2\right) \int_{z_{i \neq j}} \prod_{i \neq j} N\left(z_i ; \mu_i, \sigma_i^2\right) d z_{i \neq j} d z_i \\ & =-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J \int z_j^2 N\left(z_j ; \mu_j, \sigma_j^2\right) d z_j \\ & \left.=-\frac{J}{2} \log (2 \pi)-\frac{1}{2} \sum_{j=1}^J\left(\mu_j^2+\sigma_j^2\right)\right) \end{aligned} \]
Combining the two terms, we arrive at at the formula: \[ D_{\mathrm{KL}}\left(q_\phi(z \mid x)|| p(z)\right)=-\frac{1}{2} \sum_{j=1}^J\left(1+\log \sigma_j^2-\mu_j^2-\sigma_j^2\right) \]